Research
Published 12 November 2020At life’s edge: How genes flow in hot spring bacteria

Microbiologists have long debated how to define what a bacterial species actually is. Sexual reproduction underpins the standard concept of a biological species, but bacteria do not reproduce in this way.
Instead, they can transfer genetic material to other bacteria in a process called ‘horizontal gene transfer’. Natural bacterial communities can therefore exhibit extraordinary genomic variation within bacteria of the same species. This makes the work of microbial ecologists particularly challenging as the standard ecological theories and frameworks require the identification of genetically and ecologically cohesive populations within natural communities.
Recent advances in genomic technology and analysis have enabled microbiologists to propose novel definitions of bacterial species based on the ecological population theory. This theory describes how genes flow within and among genetically and ecologically cohesive populations of bacteria, termed ecological populations, and posits that gene-flow boundaries define bacterial species. Identifying gene-flow events, which appear as minute changes in bacterial genomes, requires very high-quality genome sequences from pure samples of single bacterial species, but these can be very difficult to obtain from natural environments. Supported by the Marsden Fund, Dr Charles Lee of the University of Waikato and his research team are using the unique geothermal microbial communities found across the national taonga of the Taupō Volcanic Zone (such as the iconic Inferno Crater Lake at Waimangu, Figure 1) to validate the ecological population theory. The team are targeting Aquificales, a taxonomic family of extraordinary bacteria that thrive in high-temperature environments and are capable of growth solely through oxidising hydrogen into water. These characteristics allow the researchers to grow these bacteria in a specific low-nutrient media in an 85% hydrogen atmosphere at a temperature of 70°C. These extreme growth conditions effectively exclude the vast majority of non-target bacteria and allow researchers to reliably isolate highly pure samples for their studies.
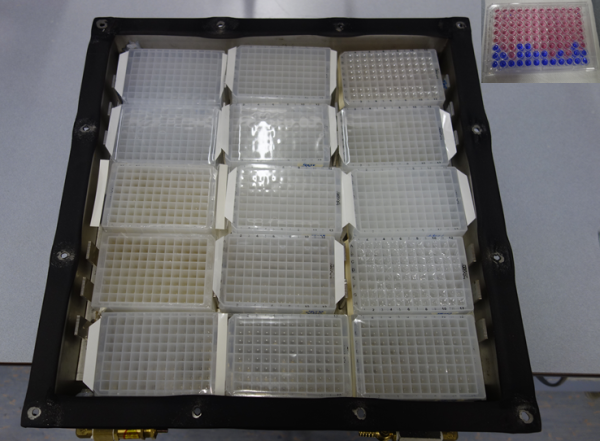
Figure 2: Growth chamber. Each column on every plate contains a dilution-to-extinction series, where the final well with growth is taken forward to the next stage to obtain pure cultures. Inset in top-right shows a resazurin plate that allows researchers to confidently determine the final well with bacterial growth by the colour change of resazurin from blue to red due to the reduction of growth media from microbial growth.
However, this unusual cultivation approach makes it extremely laborious to obtain the hundreds of isolates required for the research using conventional microbiological techniques. To overcome this, the team have designed and manufactured cost-effective and safe (hydrogen is explosive!) incubation chambers each capable of holding close to 3,000 distinct bacterial isolates (Figure 2). The team has successfully used their first-generation incubation chambers to obtain novel Aquificales genomes. Analysis of these genomic data demonstrated the existence of gene flow within Aquificales bacteria isolated from a single geothermal pool (Figure 3) and provides the first validation of the ecological population theory in a natural microbial community. Following this success, the team has improved the design and manufactured additional incubation chambers, allowing team members to study more than 20,000 isolates simultaneously across their laboratories at Waikato and Canterbury universities.
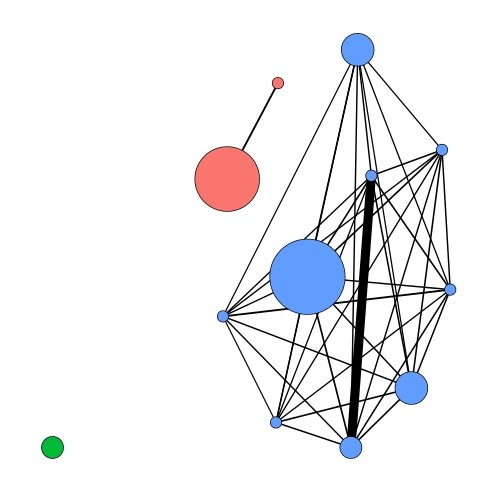
Figure 3: A network analysis of Aquificales isolates from Kuirau Park, Rotorua. Each coloured circle represents a group of Aquificales isolate genomes, with the size of the circle representing the number of genomes. Different coloured circles represent the different ecological populations identified through this analysis. The thickness of black lines represents the level of gene flow between these nodes. Note the lack of gene flow between the different coloured ecological populations.
Armed with these high-throughput capabilities for analysis of Aquificales bacteria, the team continues to conduct experiments to evaluate whether bacterial species defined by gene-flow boundaries exhibit patterns similar to those seen in sexual species. The team is also working on ways to apply their methods to other groups of bacteria and other habitats. If successful, these findings will lead to a paradigm shift in how microbial ecosystems are studied and allow the process of microbial community assembly to be studied as emergent properties of population-level interactions.
RESEARCHER
Dr Charles Lee
ORGANISATION
University of Waikato
FUNDING SUPPORT
Marsden Fund
CONTRACT OR PROJECT ID
UOW1701
